The M-sigma Relation¶
Last time, we fit a line to the M-sigma relation which related the central black hole mass to velocity dispersion of a galaxy. Today, we will try to use bootstrapping to get an estimate of the uncertainty on our fitted parameters.
First, here is the code we need from last time showing how we can fit a line to this dataset:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv('m-sigma.txt')
df.head()log_BH = np.log10(df.M_bh)
BH_err = df.M_bh_err # note: we have already converted the errors to logspace for you! fig, ax = plt.subplots(figsize=(7,7))
ax.errorbar(df.sigma, log_BH, yerr=BH_err, xerr=df.sigma_err, fmt='None', color='gray')
ax.plot(df.sigma, log_BH, 's', color='blue', ms=12, alpha=0.5, mec='k')
ax.set_xlabel('Velocity Dispersion [km/s]', fontsize=16)
ax.tick_params(which='both', top=True, right=True, direction='in', length=6)
ax.set_ylabel(r'log M$_{\rm BH}$ [$M_{\odot}$]', fontsize=16);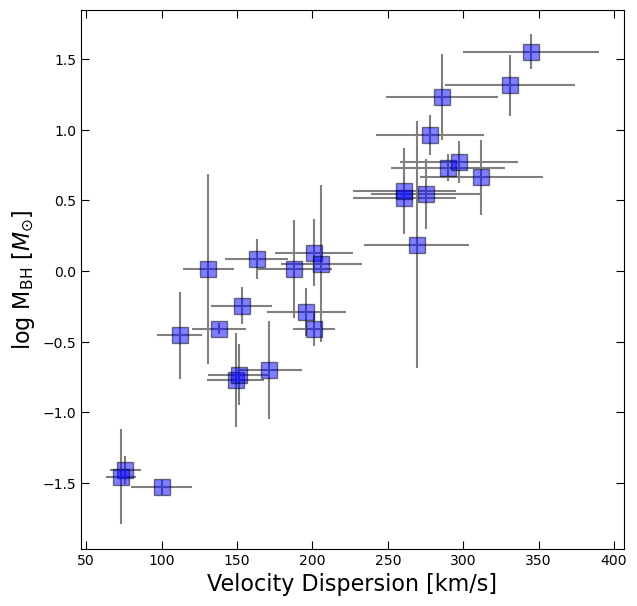
Last time, we showed you how you could use np.polyfit to fit a line. The code we used to do this looked like:
# Fit a straight line (polynomial of degree 1) to the data
coefficients = np.polyfit(x, y, deg=1)
# For a first order polynomial, there are two coefficients
# Extract the slope (m) and intercept (b) from the coefficients
slope, intercept = coefficients
# Generate points along the fitted line for plotting
x_fit = np.linspace(min(x), max(x), 100)
y_fit = slope * x_fit + interceptWe can do this more efficiently using the np.polyval function to generate y_fit without needing to write out the equation y = mx + b.
coefficients = np.polyfit(df.sigma,log_BH,deg=1,w=1/BH_err)
x_fit = np.linspace(50,400,100)
y_fit = np.polyval(coefficients, x_fit)fig, ax = plt.subplots(figsize=(7,7))
ax.errorbar(df.sigma, log_BH, yerr=BH_err, xerr=df.sigma_err, fmt='None', color='gray')
ax.plot(df.sigma, log_BH, 's', color='blue', ms=12, alpha=0.5, mec='k')
ax.plot(x_fit, y_fit, 'k', lw=3)
ax.set_xlabel('Velocity Dispersion [km/s]', fontsize=16)
ax.tick_params(which='both', top=True, right=True, direction='in', length=6)
ax.set_ylabel(r'log M$_{\rm BH}$ [$M_{\odot}$]', fontsize=16);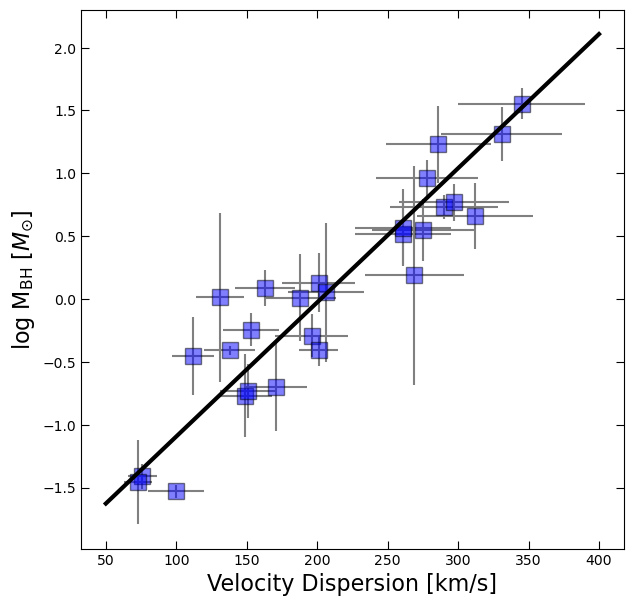
Fitting x(y)¶
We also discussed how linear least squares is only accounting for the uncertainties in the independent variable (here our -axis). But both quantities being plotted had major uncertainties, so we asked you to fit x(y) as well, using the errors on x instead of y. The solution to that attempt is below.
coeff2 = np.polyfit(log_BH,df.sigma,deg=1,w=1/df.sigma_err)
in_masses = np.linspace(-1.5,2,100)
out_velocities = np.polyval(coeff2,in_masses)fig, ax = plt.subplots(figsize=(7,7))
ax.errorbar(df.sigma, log_BH, yerr=BH_err, xerr=df.sigma_err, fmt='None', color='gray')
ax.plot(df.sigma, log_BH, 's', color='blue', ms=12, alpha=0.5, mec='k')
ax.plot(x_fit, y_fit, 'k', lw=3, label='fit y(x)')
ax.plot(out_velocities, in_masses, 'r', lw=3, label='fit x(y)')
ax.set_xlabel('Velocity Dispersion [km/s]', fontsize=16)
ax.tick_params(which='both', top=True, right=True, direction='in', length=6)
ax.legend()
ax.set_ylabel(r'log M$_{\rm BH}$ [$M_{\odot}$]', fontsize=16);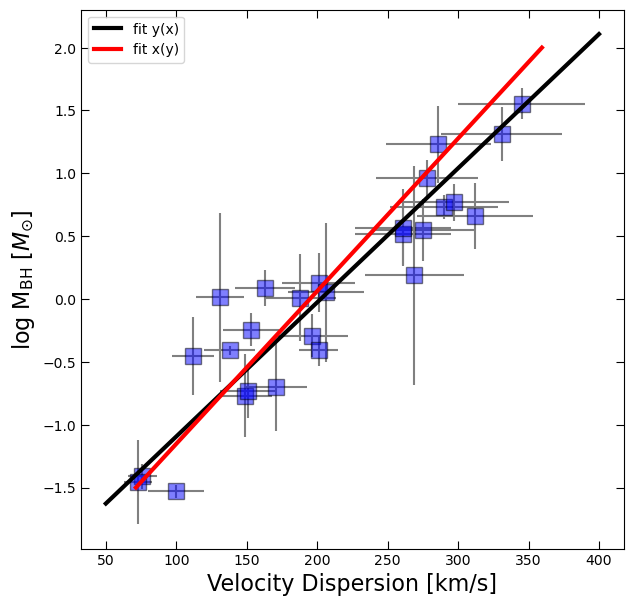
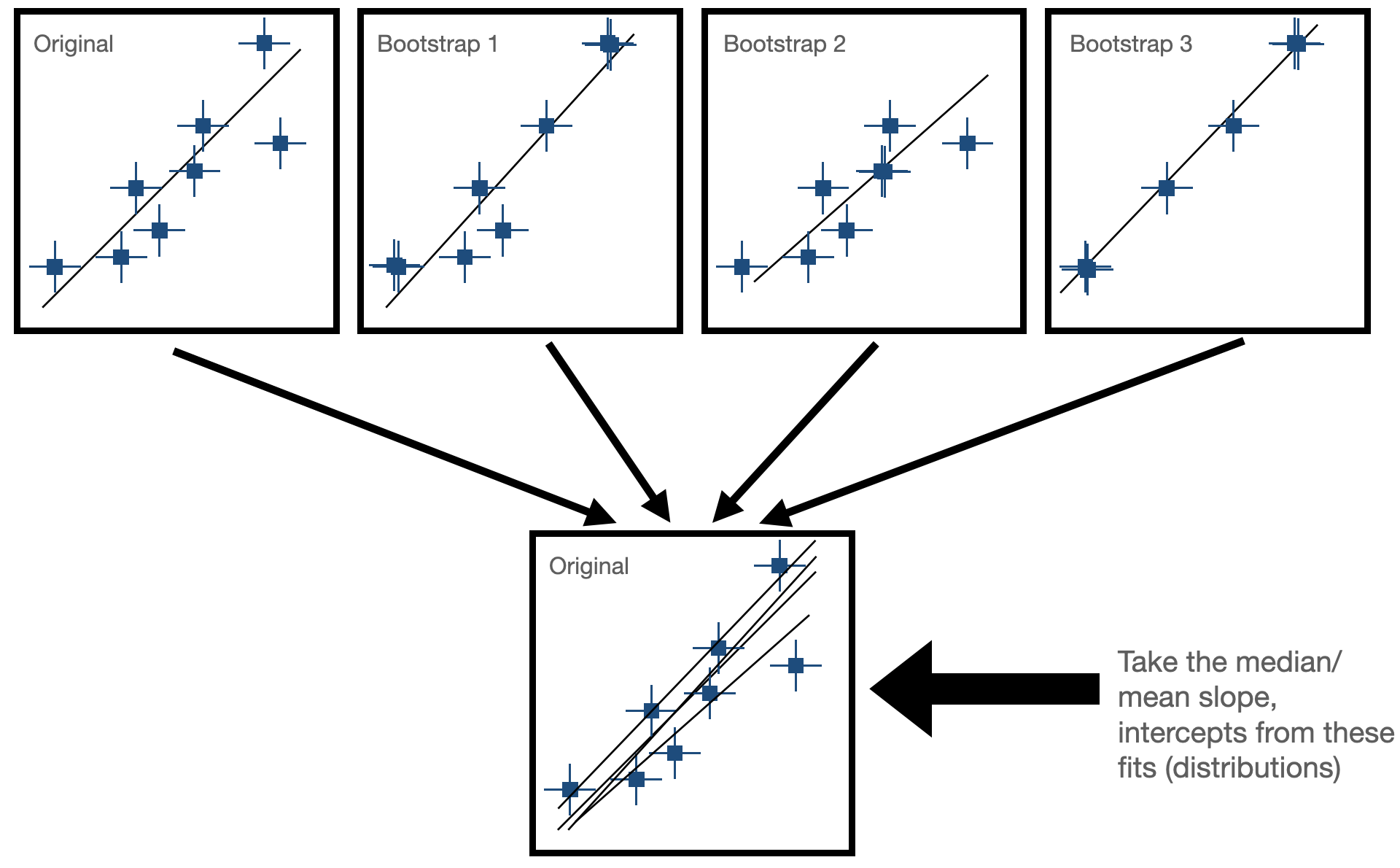
Bootstrap Resampling¶
Bootstrapping is a resampling technique used in statistics to estimate the sampling distribution of a statistic by repeatedly sampling with replacement from the observed data. It is particularly useful when the underlying population distribution is unknown or difficult to model.
- Sample Creation: Bootstrapping starts with the creation of multiple bootstrap samples by randomly selecting observations from the original dataset with replacement. This means that each observation in the original dataset has the same chance of being selected for each bootstrap sample, and some observations may be selected multiple times while others may not be selected at all.
- Statistical Estimation: After creating the bootstrap samples, the statistic of interest (e.g., mean, median, standard deviation, regression coefficient) is calculated for each bootstrap sample. This results in a distribution of the statistic across the bootstrap samples, known as the bootstrap distribution. In our case, this would be fitting our line to the data and seeing how much the paramters (slope and intercept) change for each fit. Now, we have some way of expressing uncertainty in our fitted parameters!
The goal for today is to use bootstrapping to estimate the uncertainty on our fit for the M-sigma relation!